How to collect GC-MS Spectra for GC-AutoFit
How to collect GC-MS Spectra for GC-AutoFit (GC-MS Sample Preparation Protocols)
Instruction for using User's Own Library
GC-AutoFit supports using your own library and calibration curve. In order to use this function, you must prepare your own compound library and calibration curve as per the following instructions.
Compound profiling library:
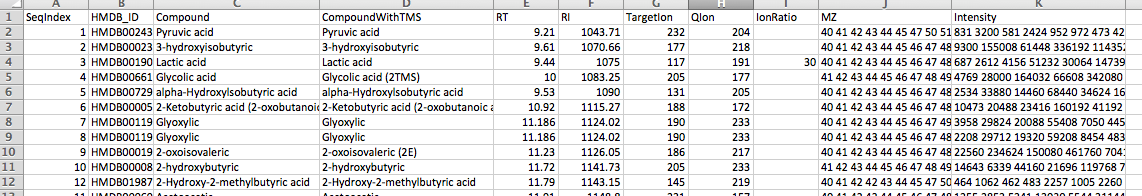
The libary file must be in CSV format and contain the following column names:
- SeqIndex
- HMDB_ID
- Compound : Compound name
- CompoundWithTMS : Compoundnd name with different TMS for detecting multiple peaks of a same compound
- RI : Retention Index
- TargetIon
- QIon
- MZ : separated with space
- Intensity : separatedd with space
Calibration curve:
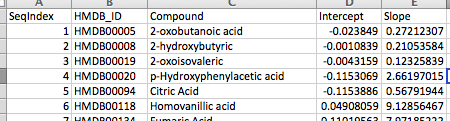
GC-AutoFit quantifies metabolite concentrations using the relative peak area and a linear model described in the calibration curve file. This file must be in CSV format and contain the following columns:
- SeqIndex
- HMDB_ID
- Compound : Compound name
- Intercept
- Slope
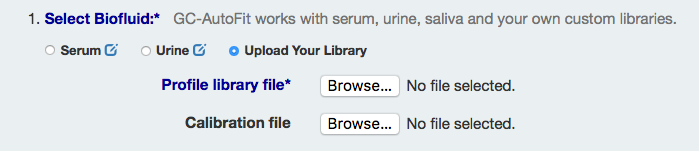
Upload User's Own Library
Choose the "Upload Your Library" in under Select Biofluid and upload your 'Profile library' and 'Calibration curve' files. The compound name of the internal standard is also required to calculate an area ratio.